Example - 74 - Uranus Aerocapture - Part 3
In this example we illustrate the selection of a equilibrium glide guidance parameters and setting up a Monte Carlo simulation for aerocapture at Uranus.
[1]:
import numpy as np
import matplotlib.pyplot as plt
from AMAT.planet import Planet
from AMAT.vehicle import Vehicle
Set up a Planet object for Uranus.
[2]:
planet = Planet('URANUS')
planet.loadAtmosphereModel('../atmdata/Uranus/uranus-gram-avg.dat', 0 , 1 ,2, 3, heightInKmFlag=True)
planet.h_skip = 1000.0E3
planet.h_low = 120e3
planet.h_trap = 100e3
Set up a vehicle object. Set the intitial state based on the discussion in example-72-uranus-aerocapture-part-2
[3]:
vehicle=Vehicle('Titania', 3000.0, 200.0 , 0.36, np.pi*4.5**2.0, 0.0, 1.125, planet)
vehicle.setInitialState(1000.0,-80.95,25.22,27.5946,132.066,-13.0 ,0.0,0.0)
vehicle.setSolverParams(1E-6)
Compute the parameters used in the equilibrium glide aerocapture guidance algorithm following Cerimele and Gamble. A Simplified Guidance Algorithm for Lifting Aeroassist Orbital Transfer Vehicles,” in 23rd Aerospace Sciences Meeting, Houston, TX, 1985, p. 348. doi: 10.2514/6.1985-348. Propogate the entry for a few seconds to compute the dynamic pressure profile during entry, and compute a linear fit: q = ah + b.
[4]:
vehicle.propogateEntry2(200, 0.1, 0)
# Values for -13.64 deg, these are the linear fit
# y = ax + b parameters used to compute Ghdot and Gq (see refs.)
a = -0.155
b = 56000
x_arr = np.linspace(260.0E3,350.0E3,101)
y_arr = a*x_arr + b
fig = plt.figure()
fig.set_size_inches([4.25,4.25])
plt.rc('font',family='Times New Roman')
params = {'mathtext.default': 'regular' }
plt.rcParams.update(params)
plt.plot(vehicle.h_kmc*1E3,vehicle.dyn_pres_atm*1.01325E5,'r-',linewidth=3.0)
plt.xlim(100.0E3, 700.0E3)
#plt.ylim(0.0,12500.0)
plt.plot(x_arr,y_arr,color='k',linewidth=2.0,linestyle='dashed')
plt.xlabel("Altitude, m", fontsize=10)
plt.ylabel("Dynamic pressure, Pa ",fontsize=10)
plt.xticks(np.array([200.0E3 ,300E3, 400.00E3, 500e3, 600.0E3]),fontsize=10)
ax=plt.gca()
ax.tick_params(direction='in')
ax.yaxis.set_ticks_position('both')
ax.xaxis.set_ticks_position('both')
ax.tick_params(axis='x',labelsize=10)
ax.tick_params(axis='y',labelsize=10)
ax.tick_params(direction='in')
ax.xaxis.set_tick_params(direction='in', which='both')
ax.yaxis.set_tick_params(direction='in', which='both')
ax.xaxis.set_tick_params(width=2, length=8)
ax.yaxis.set_tick_params(width=2, length=8)
ax.xaxis.set_tick_params(width=1, length=6, which='minor')
ax.yaxis.set_tick_params(width=1, length=6, which='minor')
ax.xaxis.grid(which='major', color='k', linestyle='dotted', linewidth=0.5)
ax.xaxis.grid(which='minor', color='k', linestyle='dotted', linewidth=0.0)
ax.yaxis.grid(which='major', color='k', linestyle='dotted', linewidth=0.5)
ax.yaxis.grid(which='minor', color='k', linestyle='dotted', linewidth=0.0)
ax.annotate(r'$\bar{q} = -0.155h + 56000$',
xy=(348586, 9103.4) ,
xytext=(431181, 9103.4),
arrowprops=dict(arrowstyle="<-"), va="center", ha="left", fontsize=9)
plt.show()
findfont: Font family ['Times New Roman'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Times New Roman'] not found. Falling back to DejaVu Sans.
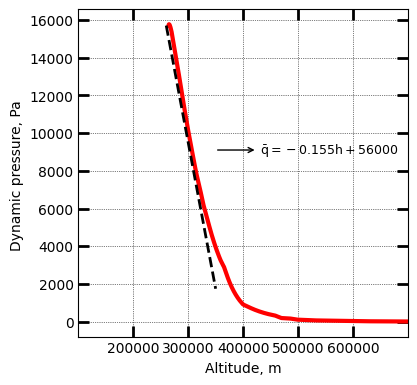
From the above, a = -0.155, b = 56000. Following Cerimel and Gamble (1985)
\begin{equation} \label{eq:omega} {\omega_n}^2 = -\frac{C_L S}{m} G_{\bar{q}} a \end{equation}
\begin{equation} \label{eq:zeta} 2 \zeta {\omega_n} = \frac{C_L S}{m} G_{\dot{h}} \end{equation}
Using \(\omega_n\) = 0.05, and \(\zeta\) = 1.5, \(G_{\bar{q}}\) and \(G_{\dot{h}}\) are computed as follows
[5]:
0.05**2*200/(0.36*0.155)
[5]:
8.960573476702512
[6]:
2*1.5*0.05*200/0.36
[6]:
83.33333333333334
The vehicle initial state (other than the EFPA) is easy to set from the approach trajectory result.
The selection of the target EFPA is a slightly tricky problem. A first estimate of the target EFPA is available from example-72-uranus-aerocapture-part-2. We assume the approach navigation is able to deliver a 1-sigma EFPA error of 0.1667 deg, or 0.50 deg (3-sigma). The EFPA must be chosen such that the (target value - 3-sigma) should be able to capture in the expected thin atmosphere (nominal density - 3-sigma), and the (target value + 3-sigma) should not result in considerable apoapsis
undershoot in case of thick atmosphere (nominal density + 3-sigma). For this example, we choose a target EFPA = -12.80 deg. This puts the -3-sigma EFPA at -12.30 deg, providing some margin (0.27 deg.) against escape in case of shallow entry combined with thin atmosphere. At the other end, the +3-sigma EFPA is -13.30 deg which also provides some margin (0.65 deg) against apoapsis undershoot in case of steep entry and thick atmosphere.
[7]:
vehicle=Vehicle('Titania', 3000.0, 200 , 0.36, np.pi*4.5**2.0, 0.0, 1.125, planet)
vehicle.setInitialState(1000.0,-80.95,25.22,27.5946,132.066,-12.80 ,0.0,0.0)
vehicle.setSolverParams(1E-6)
# Set the guidance parameters described in the paper.
# See the function description for parameter details.
# Set max roll rate constraint to 30 deg/s
vehicle.setMaxRollRate(30.0)
# Set Ghdot = 83
# Set Gq = 9.0
# Set v_switch_kms = 16.0 :: this parameter is not really used, and should just be set to low value
# Set low_Alt_km = 120
# Set numPoints_lowAlt = 101
# Set hdot_threshold = -500 m/s
vehicle.setEquilibriumGlideParams(83.0, 9.0, 16.0, 120.0, 101, -500.0)
# Set target orbit parameters
# periapsis = 4000.0 km
# apoapsis = 550,000 km :: set slightly above target (500e3) to account for bias in targeting algorithm
# apoapsis tolerance = 10 km
vehicle.setTargetOrbitParams(4000.0, 550.0E3, 10.0E3)
[8]:
# Set path to atmfiles with randomly perturbed atmosphere files.
atmfiles = ['../atmdata/Uranus/perturbed/G-01.txt',
'../atmdata/Uranus/perturbed/G-02.txt',
'../atmdata/Uranus/perturbed/G-03.txt',
'../atmdata/Uranus/perturbed/G-04.txt',
'../atmdata/Uranus/perturbed/G-05.txt']
[9]:
vehicle.setupMonteCarloSimulation(1201, 200, atmfiles, 0, 1, 2, 3, 4, True, \
-12.80, 0.1667, 0.36, 0.013, 0.5, 0.1, 2400.0)
Propogate 100 guided trajectories.
[10]:
vehicle.runMonteCarlo2(100, '../data/uranus2022/MCBX1')
RUN #: 1, SAMPLE #: 180, EFPA: -12.55, SIGMA: -0.83, LD: 0.36, APO : 524027.02
RUN #: 2, SAMPLE #: 5, EFPA: -12.68, SIGMA: 0.44, LD: 0.35, APO : 478167.79
RUN #: 3, SAMPLE #: 73, EFPA: -12.73, SIGMA: -1.82, LD: 0.37, APO : 513511.78
RUN #: 4, SAMPLE #: 77, EFPA: -12.87, SIGMA: -0.10, LD: 0.37, APO : 534446.41
RUN #: 5, SAMPLE #: 149, EFPA: -13.08, SIGMA: -0.65, LD: 0.38, APO : 490116.72
RUN #: 6, SAMPLE #: 100, EFPA: -12.83, SIGMA: -0.51, LD: 0.36, APO : 519722.91
RUN #: 7, SAMPLE #: 135, EFPA: -12.58, SIGMA: -1.75, LD: 0.37, APO : 513747.29
RUN #: 8, SAMPLE #: 4, EFPA: -12.69, SIGMA: 0.58, LD: 0.35, APO : 507983.07
RUN #: 9, SAMPLE #: 105, EFPA: -12.83, SIGMA: -1.48, LD: 0.37, APO : 503749.90
RUN #: 10, SAMPLE #: 98, EFPA: -12.73, SIGMA: -1.31, LD: 0.37, APO : 493052.87
RUN #: 11, SAMPLE #: 118, EFPA: -13.03, SIGMA: -0.68, LD: 0.37, APO : 494389.86
RUN #: 12, SAMPLE #: 119, EFPA: -12.99, SIGMA: 1.81, LD: 0.37, APO : 491178.05
RUN #: 13, SAMPLE #: 156, EFPA: -12.83, SIGMA: -0.42, LD: 0.36, APO : 524869.79
RUN #: 14, SAMPLE #: 7, EFPA: -12.82, SIGMA: 0.37, LD: 0.37, APO : 515989.66
RUN #: 15, SAMPLE #: 133, EFPA: -12.86, SIGMA: -0.53, LD: 0.36, APO : 489755.47
RUN #: 16, SAMPLE #: 176, EFPA: -12.94, SIGMA: 0.93, LD: 0.38, APO : 497603.29
RUN #: 17, SAMPLE #: 81, EFPA: -12.79, SIGMA: -1.30, LD: 0.36, APO : 481778.39
RUN #: 18, SAMPLE #: 167, EFPA: -12.76, SIGMA: 1.16, LD: 0.38, APO : 493829.19
RUN #: 19, SAMPLE #: 35, EFPA: -12.65, SIGMA: 1.51, LD: 0.37, APO : 531571.21
RUN #: 20, SAMPLE #: 168, EFPA: -12.63, SIGMA: -0.14, LD: 0.37, APO : 526488.86
RUN #: 21, SAMPLE #: 6, EFPA: -12.66, SIGMA: 0.43, LD: 0.34, APO : 520835.40
RUN #: 22, SAMPLE #: 197, EFPA: -13.04, SIGMA: 1.26, LD: 0.36, APO : 471494.05
RUN #: 23, SAMPLE #: 5, EFPA: -12.85, SIGMA: -0.04, LD: 0.36, APO : 489225.16
RUN #: 24, SAMPLE #: 188, EFPA: -12.66, SIGMA: 1.03, LD: 0.37, APO : 528314.71
RUN #: 25, SAMPLE #: 3, EFPA: -13.06, SIGMA: 0.89, LD: 0.38, APO : 524305.86
RUN #: 26, SAMPLE #: 136, EFPA: -12.54, SIGMA: 0.38, LD: 0.38, APO : 507730.05
RUN #: 27, SAMPLE #: 38, EFPA: -12.54, SIGMA: -0.57, LD: 0.37, APO : 516998.32
RUN #: 28, SAMPLE #: 79, EFPA: -12.60, SIGMA: 0.06, LD: 0.36, APO : 515393.97
RUN #: 29, SAMPLE #: 31, EFPA: -12.78, SIGMA: -1.85, LD: 0.38, APO : 530331.87
RUN #: 30, SAMPLE #: 30, EFPA: -12.52, SIGMA: 1.00, LD: 0.35, APO : 505280.49
RUN #: 31, SAMPLE #: 62, EFPA: -12.80, SIGMA: 1.06, LD: 0.35, APO : 522314.70
RUN #: 32, SAMPLE #: 120, EFPA: -12.55, SIGMA: -0.43, LD: 0.36, APO : 523096.02
RUN #: 33, SAMPLE #: 111, EFPA: -12.80, SIGMA: 1.56, LD: 0.35, APO : 495903.99
RUN #: 34, SAMPLE #: 2, EFPA: -12.71, SIGMA: -1.11, LD: 0.37, APO : 534945.17
RUN #: 35, SAMPLE #: 143, EFPA: -12.82, SIGMA: -0.03, LD: 0.34, APO : 513493.81
RUN #: 36, SAMPLE #: 105, EFPA: -12.53, SIGMA: 0.61, LD: 0.34, APO : 517309.07
RUN #: 37, SAMPLE #: 106, EFPA: -12.82, SIGMA: 0.89, LD: 0.40, APO : 488196.50
RUN #: 38, SAMPLE #: 25, EFPA: -12.90, SIGMA: -0.37, LD: 0.36, APO : 545628.94
RUN #: 39, SAMPLE #: 124, EFPA: -12.80, SIGMA: 1.42, LD: 0.37, APO : 499014.22
RUN #: 40, SAMPLE #: 137, EFPA: -12.70, SIGMA: 0.81, LD: 0.34, APO : 512430.75
RUN #: 41, SAMPLE #: 34, EFPA: -12.69, SIGMA: 0.45, LD: 0.34, APO : 510861.72
RUN #: 42, SAMPLE #: 77, EFPA: -12.75, SIGMA: 0.03, LD: 0.35, APO : 522768.12
RUN #: 43, SAMPLE #: 74, EFPA: -12.56, SIGMA: -1.05, LD: 0.35, APO : 551946.45
RUN #: 44, SAMPLE #: 39, EFPA: -12.96, SIGMA: -0.18, LD: 0.37, APO : 533457.26
RUN #: 45, SAMPLE #: 70, EFPA: -12.70, SIGMA: -0.27, LD: 0.35, APO : 497373.61
RUN #: 46, SAMPLE #: 108, EFPA: -12.91, SIGMA: 1.25, LD: 0.35, APO : 484137.24
RUN #: 47, SAMPLE #: 18, EFPA: -13.05, SIGMA: 0.37, LD: 0.35, APO : 476890.11
RUN #: 48, SAMPLE #: 2, EFPA: -12.80, SIGMA: -0.17, LD: 0.36, APO : 541290.54
RUN #: 49, SAMPLE #: 14, EFPA: -12.86, SIGMA: -2.28, LD: 0.36, APO : 487840.40
RUN #: 50, SAMPLE #: 58, EFPA: -12.82, SIGMA: -0.88, LD: 0.36, APO : 503981.29
RUN #: 51, SAMPLE #: 113, EFPA: -12.56, SIGMA: -0.47, LD: 0.34, APO : 523246.70
RUN #: 52, SAMPLE #: 171, EFPA: -12.85, SIGMA: -1.26, LD: 0.35, APO : 504941.78
RUN #: 53, SAMPLE #: 17, EFPA: -12.81, SIGMA: 1.02, LD: 0.36, APO : 492121.66
RUN #: 54, SAMPLE #: 79, EFPA: -13.00, SIGMA: 0.84, LD: 0.36, APO : 479304.88
RUN #: 55, SAMPLE #: 134, EFPA: -12.66, SIGMA: -0.17, LD: 0.37, APO : 493614.90
RUN #: 56, SAMPLE #: 115, EFPA: -13.11, SIGMA: 0.55, LD: 0.37, APO : 484755.64
RUN #: 57, SAMPLE #: 118, EFPA: -12.48, SIGMA: -0.38, LD: 0.34, APO : 531621.18
RUN #: 58, SAMPLE #: 20, EFPA: -12.86, SIGMA: -0.03, LD: 0.36, APO : 485232.59
RUN #: 59, SAMPLE #: 130, EFPA: -12.98, SIGMA: 0.25, LD: 0.36, APO : 490032.26
RUN #: 60, SAMPLE #: 141, EFPA: -12.48, SIGMA: 0.37, LD: 0.36, APO : 516582.02
RUN #: 61, SAMPLE #: 169, EFPA: -12.84, SIGMA: 1.22, LD: 0.33, APO : 500604.74
RUN #: 62, SAMPLE #: 118, EFPA: -12.41, SIGMA: 1.16, LD: 0.35, APO : 525646.86
RUN #: 63, SAMPLE #: 179, EFPA: -13.12, SIGMA: 0.37, LD: 0.35, APO : 466605.95
RUN #: 64, SAMPLE #: 181, EFPA: -12.37, SIGMA: -0.97, LD: 0.35, APO : 558141.63
RUN #: 65, SAMPLE #: 53, EFPA: -12.65, SIGMA: 0.24, LD: 0.37, APO : 512587.08
RUN #: 66, SAMPLE #: 175, EFPA: -12.72, SIGMA: 1.14, LD: 0.34, APO : 501872.14
RUN #: 67, SAMPLE #: 169, EFPA: -13.04, SIGMA: -0.17, LD: 0.39, APO : 495872.18
RUN #: 68, SAMPLE #: 20, EFPA: -12.83, SIGMA: 0.46, LD: 0.35, APO : 521723.34
RUN #: 69, SAMPLE #: 174, EFPA: -12.76, SIGMA: -1.63, LD: 0.37, APO : 487852.34
RUN #: 70, SAMPLE #: 43, EFPA: -12.58, SIGMA: 0.64, LD: 0.35, APO : 497722.47
RUN #: 71, SAMPLE #: 53, EFPA: -12.63, SIGMA: 0.41, LD: 0.35, APO : 503835.13
RUN #: 72, SAMPLE #: 110, EFPA: -12.68, SIGMA: 0.26, LD: 0.36, APO : 532383.09
RUN #: 73, SAMPLE #: 142, EFPA: -12.70, SIGMA: -0.81, LD: 0.35, APO : 515436.90
RUN #: 74, SAMPLE #: 66, EFPA: -12.65, SIGMA: -1.08, LD: 0.35, APO : 527546.06
RUN #: 75, SAMPLE #: 142, EFPA: -12.73, SIGMA: -0.31, LD: 0.37, APO : 515880.01
RUN #: 76, SAMPLE #: 103, EFPA: -12.85, SIGMA: -0.55, LD: 0.36, APO : 494269.99
RUN #: 77, SAMPLE #: 9, EFPA: -12.82, SIGMA: -1.38, LD: 0.36, APO : 507053.59
RUN #: 78, SAMPLE #: 71, EFPA: -12.92, SIGMA: -0.68, LD: 0.38, APO : 496068.24
RUN #: 79, SAMPLE #: 30, EFPA: -12.76, SIGMA: -0.41, LD: 0.36, APO : 524483.08
RUN #: 80, SAMPLE #: 175, EFPA: -12.85, SIGMA: 0.73, LD: 0.36, APO : 500812.75
RUN #: 81, SAMPLE #: 12, EFPA: -12.81, SIGMA: -0.29, LD: 0.34, APO : 527159.37
RUN #: 82, SAMPLE #: 98, EFPA: -12.66, SIGMA: 0.36, LD: 0.35, APO : 520884.95
RUN #: 83, SAMPLE #: 42, EFPA: -12.95, SIGMA: 0.18, LD: 0.37, APO : 486903.20
RUN #: 84, SAMPLE #: 139, EFPA: -12.66, SIGMA: -1.89, LD: 0.37, APO : 518124.02
RUN #: 85, SAMPLE #: 42, EFPA: -12.98, SIGMA: 1.05, LD: 0.34, APO : 494035.94
RUN #: 86, SAMPLE #: 159, EFPA: -12.86, SIGMA: -0.62, LD: 0.35, APO : 477250.88
RUN #: 87, SAMPLE #: 108, EFPA: -12.71, SIGMA: 2.01, LD: 0.35, APO : 485696.98
RUN #: 88, SAMPLE #: 52, EFPA: -12.63, SIGMA: -0.30, LD: 0.35, APO : 528134.82
RUN #: 89, SAMPLE #: 195, EFPA: -12.60, SIGMA: 0.40, LD: 0.36, APO : 502138.06
RUN #: 90, SAMPLE #: 128, EFPA: -12.91, SIGMA: -0.39, LD: 0.35, APO : 519437.38
RUN #: 91, SAMPLE #: 71, EFPA: -12.83, SIGMA: -2.33, LD: 0.35, APO : 531887.18
RUN #: 92, SAMPLE #: 188, EFPA: -12.83, SIGMA: 0.07, LD: 0.35, APO : 494853.64
RUN #: 93, SAMPLE #: 163, EFPA: -12.55, SIGMA: -1.53, LD: 0.35, APO : 521681.44
RUN #: 94, SAMPLE #: 163, EFPA: -12.49, SIGMA: 0.15, LD: 0.35, APO : 533140.78
RUN #: 95, SAMPLE #: 187, EFPA: -12.68, SIGMA: -0.46, LD: 0.37, APO : 508596.95
RUN #: 96, SAMPLE #: 188, EFPA: -12.64, SIGMA: 1.70, LD: 0.37, APO : 505111.16
RUN #: 97, SAMPLE #: 124, EFPA: -12.72, SIGMA: 0.98, LD: 0.35, APO : 521911.31
RUN #: 98, SAMPLE #: 142, EFPA: -12.72, SIGMA: 0.99, LD: 0.32, APO : 537695.15
RUN #: 99, SAMPLE #: 181, EFPA: -12.53, SIGMA: -0.89, LD: 0.37, APO : 527053.16
RUN #: 100, SAMPLE #: 87, EFPA: -12.60, SIGMA: 0.60, LD: 0.37, APO : 525199.55
[12]:
p1 = np.loadtxt('../data/uranus2022/MCBX1/terminal_periapsis_arr.txt')
x1 = np.loadtxt('../data/uranus2022/MCBX1/terminal_apoapsis_arr.txt')
y1 = np.loadtxt('../data/uranus2022/MCBX1/acc_net_g_max_arr.txt')
z1 = np.loadtxt('../data/uranus2022/MCBX1/q_stag_max_arr.txt')
h1 = np.loadtxt('../data/uranus2022/MCBX1/heatload_max_arr.txt')
u1 = np.loadtxt('../data/uranus2022/MCBX1/periapsis_raise_DV_arr.txt')
v1 = np.loadtxt('../data/uranus2022/MCBX1/apoapsis_raise_DV_arr.txt')
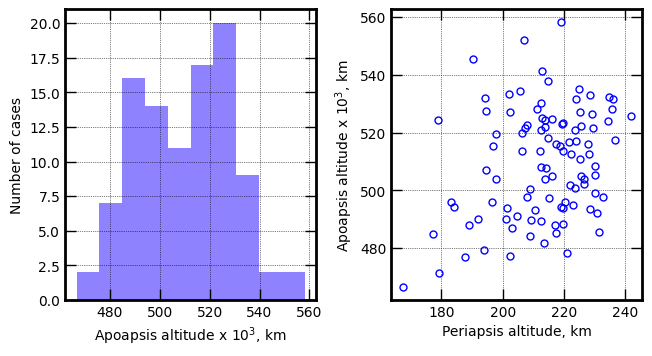
Plot apoapsis historgram and apoapsis vs periapsis (before periapsis raise maneuver)
[23]:
fig, axes = plt.subplots(1, 2, figsize = (6.5, 3.5))
fig.tight_layout()
plt.subplots_adjust(wspace=0.30)
axes[0].hist(x1/1000.0, bins=10, color='xkcd:periwinkle')
axes[0].set_xlabel('Apoapsis altitude x '+r'$10^3$'+', km', fontsize=10)
axes[0].set_ylabel('Number of cases', fontsize=10)
axes[0].tick_params(direction='in')
axes[0].yaxis.set_ticks_position('both')
axes[0].xaxis.set_ticks_position('both')
axes[0].tick_params(axis='x',labelsize=10)
axes[0].tick_params(axis='y',labelsize=10)
axes[1].plot(p1, x1/1000.0, 'bo', markersize=5, mfc='None')
axes[1].set_xlabel('Periapsis altitude, km',fontsize=10)
axes[1].set_ylabel('Apoapsis altitude x '+r'$10^3$'+', km', fontsize=10)
axes[1].tick_params(direction='in')
axes[1].yaxis.set_ticks_position('both')
axes[1].xaxis.set_ticks_position('both')
axes[1].tick_params(axis='x',labelsize=10)
axes[1].tick_params(axis='y',labelsize=10)
for ax in axes:
ax.xaxis.set_tick_params(direction='in', which='both')
ax.yaxis.set_tick_params(direction='in', which='both')
ax.xaxis.set_tick_params(width=1, length=8)
ax.yaxis.set_tick_params(width=1, length=8)
ax.xaxis.set_tick_params(width=1, length=6, which='minor')
ax.yaxis.set_tick_params(width=1, length=6, which='minor')
ax.xaxis.grid(which='major', color='k', linestyle='dotted', linewidth=0.5)
ax.xaxis.grid(which='minor', color='k', linestyle='dotted', linewidth=0.0)
ax.yaxis.grid(which='major', color='k', linestyle='dotted', linewidth=0.5)
ax.yaxis.grid(which='minor', color='k', linestyle='dotted', linewidth=0.0)
for axis in ['top', 'bottom', 'left', 'right']:
ax.spines[axis].set_linewidth(2)
plt.show()
[25]:
fig, axes = plt.subplots(1, 2, figsize = (6.5, 3.5))
fig.tight_layout()
plt.subplots_adjust(wspace=0.30)
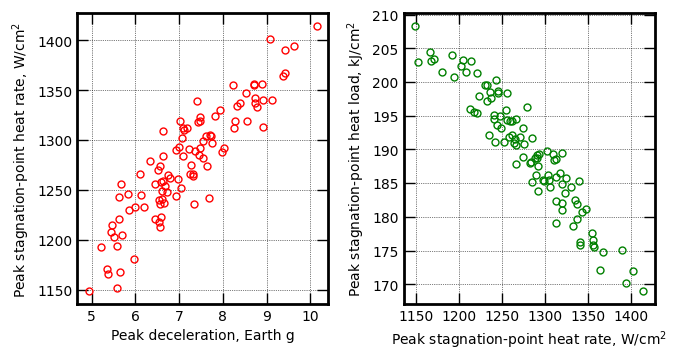
axes[0].plot(y1, z1, 'ro', markersize=5, mfc='None')
axes[0].set_xlabel('Peak deceleration, Earth g', fontsize=10)
axes[0].set_ylabel('Peak stagnation-point heat rate, '+r'$W/cm^2$', fontsize=10)
axes[0].tick_params(direction='in')
axes[0].yaxis.set_ticks_position('both')
axes[0].xaxis.set_ticks_position('both')
axes[0].tick_params(axis='x',labelsize=10)
axes[0].tick_params(axis='y',labelsize=10)
axes[1].plot(z1, h1, 'go', markersize=5, mfc='None')
axes[1].set_xlabel('Peak stagnation-point heat rate, '+r'$W/cm^2$', fontsize=10)
axes[1].set_ylabel('Peak stagnation-point heat load, '+r'$kJ/cm^2$', fontsize=10)
axes[1].tick_params(direction='in')
axes[1].yaxis.set_ticks_position('both')
axes[1].xaxis.set_ticks_position('both')
axes[1].tick_params(axis='x',labelsize=10)
axes[1].tick_params(axis='y',labelsize=10)
for ax in axes:
ax.xaxis.set_tick_params(direction='in', which='both')
ax.yaxis.set_tick_params(direction='in', which='both')
ax.xaxis.set_tick_params(width=1, length=8)
ax.yaxis.set_tick_params(width=1, length=8)
ax.xaxis.set_tick_params(width=1, length=6, which='minor')
ax.yaxis.set_tick_params(width=1, length=6, which='minor')
ax.xaxis.grid(which='major', color='k', linestyle='dotted', linewidth=0.5)
ax.xaxis.grid(which='minor', color='k', linestyle='dotted', linewidth=0.0)
ax.yaxis.grid(which='major', color='k', linestyle='dotted', linewidth=0.5)
ax.yaxis.grid(which='minor', color='k', linestyle='dotted', linewidth=0.0)
for axis in ['top', 'bottom', 'left', 'right']:
ax.spines[axis].set_linewidth(2)
plt.show()
[30]:
fig, axes = plt.subplots(1, 2, figsize = (6.5, 3.5))
fig.tight_layout()
plt.subplots_adjust(wspace=0.30)
axes[0].hist(u1, bins=10, color='xkcd:periwinkle')
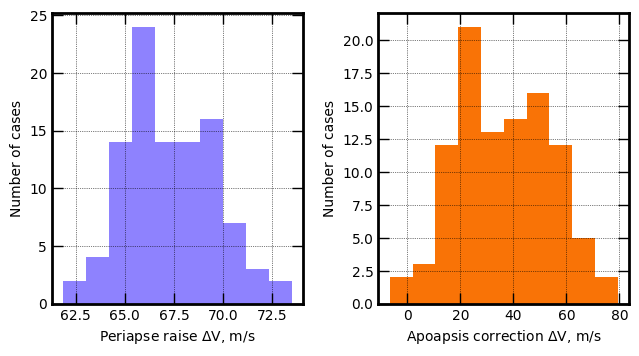
axes[0].set_xlabel('Periapse raise '+r'$\Delta V$'+', m/s', fontsize=10)
axes[0].set_ylabel('Number of cases', fontsize=10)
axes[0].tick_params(direction='in')
axes[0].yaxis.set_ticks_position('both')
axes[0].xaxis.set_ticks_position('both')
axes[0].tick_params(axis='x',labelsize=10)
axes[0].tick_params(axis='y',labelsize=10)
axes[1].hist(v1, bins=10, color='xkcd:orange')
axes[1].set_xlabel('Apoapsis correction '+r'$\Delta V$'+', m/s', fontsize=10)
axes[1].set_ylabel('Number of cases', fontsize=10)
axes[1].tick_params(direction='in')
axes[1].yaxis.set_ticks_position('both')
axes[1].xaxis.set_ticks_position('both')
axes[1].tick_params(axis='x',labelsize=10)
axes[1].tick_params(axis='y',labelsize=10)
for ax in axes:
ax.xaxis.set_tick_params(direction='in', which='both')
ax.yaxis.set_tick_params(direction='in', which='both')
ax.xaxis.set_tick_params(width=1, length=8)
ax.yaxis.set_tick_params(width=1, length=8)
ax.xaxis.set_tick_params(width=1, length=6, which='minor')
ax.yaxis.set_tick_params(width=1, length=6, which='minor')
ax.xaxis.grid(which='major', color='k', linestyle='dotted', linewidth=0.5)
ax.xaxis.grid(which='minor', color='k', linestyle='dotted', linewidth=0.0)
ax.yaxis.grid(which='major', color='k', linestyle='dotted', linewidth=0.5)
ax.yaxis.grid(which='minor', color='k', linestyle='dotted', linewidth=0.0)
for axis in ['top', 'bottom', 'left', 'right']:
ax.spines[axis].set_linewidth(2)
plt.show()